Regulatory module update in human and mouse cisRED databases
Regulatory module predictions
Translating whole-genome sets of significant atomic motifs into putative regulatory modules requires an intermediate step - identifying groups of similar motifs; modules are co-occurring group labels that satisfy certain criteria. We identify groups of similar motifs by a) annotating atomic motifs with known binding site resources from TRANSFAC, JASPAR or ORegAnno; and b) de novo motif clustering. A pattern or module is either de novo, annotation-based or hybrid, depending on the type(s) of motif groups it contains.
While the human 8 and mouse 2 databases offered de novo patterns, the human 8.1 and mouse 2.1 databases now offer both annotation-based and de novo patterns. Currently we consider annotation-based groups and patterns to be more reliable than 'de novo' groups and patterns. We are working to improve genome-scale de novo grouping.
Genome browser display
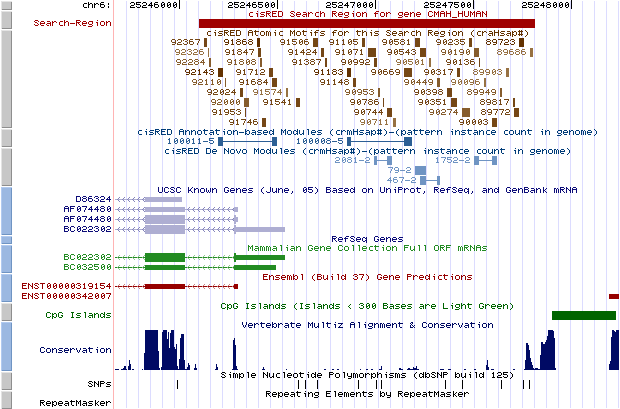
To facilitate interpreting the cisRED data, each Gene page now shows a UCSC genome browser view that gives the annotation context of a cisRED search region, significant conserved motifs and predicted regulatory modules. Clicking on the screenshot displays that gene's cisRED annotations in a live session at the UCSC genome browser.
In the genome browser view, the horizontal red bar at the top shows the nominal search region within which comparative genomics discovery methods were applied outside of coding exons and most types of repeats. The numbered brown blocks are atomic motifs, i.e. conserved DNA sequence motifs that were identified by discovery methods and post-processing operations. Motifs are shaded to indicate the discovery p-value; a darker motif was more significant at the discovery stage. Following motif discovery, motifs were filtered by membership in co-occurring patterns, and patterns were ranked by genome-scale properties. Motifs instances that occur in highly ranked putative regulatory modules may be more reliable predictions of functional genomic elements. The connected sets of blue boxes are co-occurring patterns, i.e. putative regulatory modules. Modules are shaded to indicate the number of times that a pattern is found in the target genome; a darker module is associated with more search regions.