Cryptococcus Neoformans Summary
Whole-Genome Shotgun Sequence of a Human Pathogen
With funding from Genome BC and Genome Canada, the Genome Sciences Centre is sequencing the genome of strain WM276 of the fungal pathogen C. neoformans as part of a collaboration with Jim Kronstad in the Biotechnology Laboratory at the University of British Columbia. C. neoformans causes meningoencephalitis in humans and animals Strains of the fungus exist in five serotypes (A, B, C, D and AD) and the WM276 strain represents the serotype B group. In general, cryptococcal infections occur at high frequency (10 - 25%) in immunocompromised people such as those individuals infected with the human immunodeficiency virus (HIV). However, serotype B strains also infect immunocompetent (otherwise healthy) people as demonstrated by the recent outbreak of infections caused by the fungus on Vancouver Island. The genome sequence of the serotype B strain WM276 will provide important information to help with the identification of cryptococcal strains from Vancouver Island and around the world. It will also provide important comparative information to identify genes in the fungus that may play a role in disease. In addition, the sequence will be a valuable resource for developing diagnostic tests to classify strains with regard to serotype and to identify potential targets for antifungal therapy.

Cryptococcus cells labeled with green fluorescent protein
The genome sequencing project for strain WM276 is one component of a larger genomics program for C. neoformans that involves additional collaborations with Jim Kronstad and several genome centres around the world. Specifically, we have constructed physical maps of fingerprinted Bacterial Artificial Chromosome (BAC) clones for each of the four strains of C. neoformans that are being used in genome sequencing projects. These projects are underway at Stanford University (Stanford Genome Technology Center), Duke University (Center for Genome Technology), The Institute for Genomic Research and the Whitehead Institute (Center for Genome Research). The maps provide valuable resources for the assembly of whole genome sequencing data and they aid in the completion of the genomes. In addition, the maps allow rapid comparison of the genomes of different serotypes (Schein et al.,2002). Finally, we are also characterizing the transcriptome of serotype A, B and D strains of C. neoformans using serial analysis of gene expression (SAGE; Steen et al.2002; 2003).
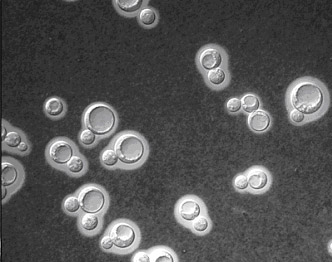
Cryptococcus cells with polysaccharide capsule
Data Release Policy (for use with sequence access)
With funding from Genome Canada, we have initiated a project to complete the genomic sequence of the Cryptococcus neoformans strain WM276 (serotype B). Our intention is to publish the genome sequence as soon as possible in a peer reviewed journal, with an emphasis on comparative studies with other completed C. neoformans genomes. The data are freely available, although users should contact Jim Kronstad if they intend to perform large-scale analyses (e.g., chromosome or whole genome studies). The data should be regarded as preliminary and subject to change as the project proceeds. We request that users of the data reference the following citation: "These sequence data were produced at Canada's Michael Smith Genome Sciences Centre with funding from Genome Canada and are available at www.bcgsc.ca.
Research Team
Jim Kronstad, Cletus D'Souza, Marco Marra, Steven Jones, Rob Holt, René Warren, George Yang, Yaron Butterfield, Jeff Stott, Asim Siddiqui
