Forestry Genomics Summary
Poplar - Industrially Important Pulp Source
![]() In collaboration with Kermit Ritland
(UBC, Forest Sciences),
Jorg Bohlmann
(UBC, Biotech Lab/Forest Sciences/Botany),
Carl Douglas (UBC, Botany),
Brian Ellis
(UBC, Biotech Lab/Ag. Sciences), this project involves mapping the poplar genome as well as carrying
out BAC end sequencing to add to the utility of the map. The poplar genome will add to the numerous
mamallian and other genomes already mapped by the Genome Sciences Centre.
In collaboration with Kermit Ritland
(UBC, Forest Sciences),
Jorg Bohlmann
(UBC, Biotech Lab/Forest Sciences/Botany),
Carl Douglas (UBC, Botany),
Brian Ellis
(UBC, Biotech Lab/Ag. Sciences), this project involves mapping the poplar genome as well as carrying
out BAC end sequencing to add to the utility of the map. The poplar genome will add to the numerous
mamallian and other genomes already mapped by the Genome Sciences Centre.
The goal of this proposal is to maximize the value of Canadian forests by unlocking the genetic potential of two important species (spruce and poplar) and focusing on two areas critical to the future of Canadian forestry, forest health and wood quality. Species of spruce (Picea) are the dominant genera of the boreal forest and occur throughout Canada. Five species occur in Canada: Black (Picea mariana), red (Picea rubens), white (Picea glauca), Engelmann (Picea engelmannii) and Sitka (Picea sitchensis). Norway spruce (Picea abies) is also a valuable species in Northern Europe. In the forest industry, spruce is the predominant harvested tree species for both structural timber and pulp. All species in Canada are the subject of conventional tree breeding programs for growth, wood quality and pest resistance. Aspen (Populus tremuloides) is the most widespread hardwood in Canada. Other important members of the genus in Canada include P. balsamifera and P. deltoides. Poplar breeding and selection programs in several provenances in Canada and the US have provided a large base of selected and pedigreed material. "Hybrid" poplar, or progeny of crosses between species, often shows exceptional vigor and growth, and is typically grown in plantations. This proposal focuses on white and black spruce, and on trembling aspen. In addition, hybrids and backcrosses with close relatives of these species will be used.
The goal of this proposal is to maximize the value of Canadian forests by unlocking the genetic potential of two important species (spruce and poplar) and focusing on two areas critical to the future of Canadian forestry, forest health and wood quality.
Our proposal also includes comparisons with Arabidopsis, whose complete genome was recently sequenced Spruce and poplar (Populus spp.) have complementary characteristics that facilitate an integrated forestry genomics project. Poplar is a "model tree" species, while spruce represents a phylum (conifers) that dominates the world's temperate and boreal forests and provides Canada with high quality wood and fibre. The large genome of spruce mandates more functional approaches, while the small genome of poplar makes structural approaches possible. The phylogenetic distance between spruce and angiosperms makes evolutionary comparisons of spruce sequences informative. The small genome of poplar makes construction of integrated physical and genetic maps feasible, and its relative closeness to species such as Arabidopsis makes the large-scale conservation of gene order (synteny) more likely. Spruce is economically dominant in the Canadian forest industry, but as forestry shifts towards more intensive cultivation in smaller areas, poplar may become increasingly important as a short-rotation, highly productive species.
To provide a broader perspective on the relationship of spruce and poplar to other vascular plant species, below is given a phylogeny depicting evolutionary relationships of these two species among representative gymnosperms and angiosperms.
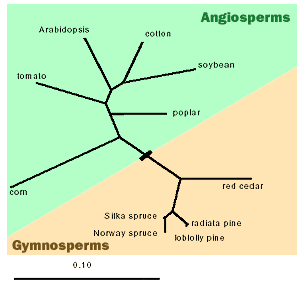
Only those angiosperms currently subject to genome projects are given (the phylogeny is based on the chloroplast rbcL gene). Gymnosperms and angiosperms are thought to have diverged approximately 160 - 200 million years ago, and Populus and Arabidopsis about 50 million years ago. Though phylogenetically distantly related, angiosperms and gymnosperms are both members of the monophyletic group, the Tracheophyta, which are land plants containing a vascular system characterized by the presence of tracheary elements (TE) of the xylem. TE cells are the major constituents of secondary xylem, or wood. Although the woody growth habit has evolved multiple times in different angiosperm lineages (families), it is likely that the mechanisms dictating vascular system development and TE differentiation are conserved among vascular plants, and that most genes controlling wood formation and wood quality are homologous across all these species. By contrast, it is likely that spruce, poplar and Arabidopsis have evolved unique mechanisms for defense against insects and disease -- it is well known that defense related genes evolve quickly in response to changing biotic pressures. Thus, homology-based approaches using information from other model species are less appropriate for probing the nature of spruce defense mechanisms.
