Full-Genome Human BAC Rearrays
Human genome high-resolution BAC rearray set
| Annotations |
|---|
|
Contact: Martin Krzywinski (BCGSC) |
| Browse Clone Set |
|---|
|
A track showing the localized clones in the rearray is available through the UCSC Genome Browser. |
| Distribution |
|---|
|
Information about distribution can be found at the BACPAC Resource Centre pages, or contact Pieter de Jong (BACPAC) or Kazutoyo Osoegawa (BACPAC) |
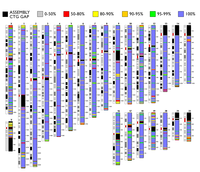
The BAC clone set for Homo sapiens is comprised of 32,432 BAC clones. The set has an average resolution of 45 kb (average clone cover size) and an effective resolution of 75 kb (weighted clone cover size). The set provides coverage for 99% of both the fingerprint map and the current sequence assembly. The clones were selected using the November 2001 fingerprint map from Washington University Genome Sequencing Centre and the August 2001 hg8 human genome assembly from UCSC.
All data available here is based the analysis of the clone set using the April 2003 hg15 UCSC sequence assembly and June 2003 human fingerprint map. Currently, there are 32 Mb in 613 gaps in coverage of the sequence assembly with 25% of the gaps being smaller than 10 kb. The clones have been selected from the fingerprint map to provide maximum representation of every clone in the map. Additional clones were added to the set to cover regions which were thought to lack coverage. For additional information please contact Martin Krzywinski (BCGSC).
The BAC clones are distributed through Pieter de Jong's BACPAC Resource Centre. The set exists as a 32,432 clone distribution as well as chromosome-specific sub-sets. Information about the distribution and cost of the set is available at BACPAC. To obtain more information about ordering the rearrayed BACs contact Pieter de Jong (BACPAC) or Kazutoyo Osoegawa (BACPAC).
Details about the set and the selection process have been presented in a poster at the 2003 Advances in Genome Biology and Technology Conference at Marco Island, Florida (download PDF poster).
download PDF poster | AGBT Abstract
A Set of Rearrayed BAC Clones spanning the human genome
Krzywinski M, Bosdet I, Smailus D, Mathewson C, Wye N, Barber S, Brown-John M, Chand S, Cloutier A Masson A, Mayo M, Olson T, Lam W, MacAuley C, Osoegawa K†, Zhao S‡, de Jong PJ†, Schein J, Jones S, Marra M
†Children’s Hospital Oakland Research Institute, Oakland, CA, USA
‡The Institute for Genomic Research, Rockville, MD, USA
From the human fingerprint map constructed at Washington University Genome Sequencing Center, we have selected a set of 32,433 BACs that span the human genome. The purpose of the clone set is to serve as a genome-ordered set of probes for FISH and microarray-based BAC CGH experiments. The comprehensive coverage of this clone set makes it a valuable asset in both research and clinical contexts, in the search for understanding and detection of cancer-related chromosomal and expression alterations.
The clones have been sampled from RPCI-11 and RPCI-13 (94%) and Caltech-D (6%) libraries, selected to optimize size, coverage of the map and consistent overlap. The clones have been rearrayed into 384-well format. The identity of clones has been validated by fingerprinting. Following the first round selection of 29,035 clones, a combination of automated and visual fingerprint inspection identified 1,978 clones that did not match the fingerprints stored in the fingerprint map. 4,531 clones were added to the set to maximally conserve map coverage of the unmatched clones. Analysis of the set's sequence coverage (UCSC, 2002/06 assembly) resulted in the selection of an additional 1,258 clones, with some chosen from outside the fingerprint map, to cover gaps larger than 10 kb. During the second round of fingerprint validation 413 clones were rejected.
The clone set covers 99% of the November 2001 version of the BAC fingerprint map. Using fingerprint-based localization, end sequence data and assembly coordinate data, 30,561 of the clones were localized within the genome and found to cover 2.788 Gb (99%+) of the assembled sequence. Approximately 35 Mb of this coverage was provided by clones not found in the fingerprint map. Approximately 82% of the assembly is covered at 1X and 2X in a 1:1 ratio. The sequence coverage of the set contains 729 sequence coverage gaps totaling 24Mb, with 46% of the gaps being smaller than 10kb. The average resolution of the clone set is 46kb.
This first version of the clone set is publicly distributed through the BACPAC Resources Centre (Children’s Hospital, Oakland). We anticipate that the set will evolve as new versions of the sequence assembly and physical map are released. We are planning to create an analogous resource for the mouse and rat genomes.
Annotations
lists
The annotations were produced using the July 2002 UCSC sequence assembly and the November 2001 fingerprint map.
master clone list
This file contains the master clone list of the rearray set. Each clone is
reported by its mapping name, Genbank name and FPC name.
clones and locations
This list provides the names and genome locations for the clones in the set. The naming convention is D/M for CTD clones, N for RP11 and F for RP13. Thus N0123A01 is R11-123A1. The list contains the following information
- clonename
- canonical name
- fingerprint map contig
- coordinate type
- chromosome
- start position
- end position
- accession
- sequence status
- chromosome assignment in Genbank
The coordinate type field indicates the type of sequence coordinate reported in the file for each clone.
- BESxyy BES or Golden Path coordinate which overlaps with fingerprint-based
coordinates by yy %.
- x = "V" if middle(BES) - middle(ANC) < 10kb
- x = "v" if middle(BES) - middle(ANC) < 20kb
- x = "w" if middle(BES) - middle(ANC) < 50kb
- x = "f" if middle(BES) - middle(ANC) < 100kb
- x = "F" if middle(BES) - middle(ANC) >= 100kb
- y = "++" if the BES/ANC overlap is 100%
- y = "--" if there is no BES/ANC overlap
- xyy = "DAN" if the BES/ANC coordinates are on different neighbourhoods
- xyy = "INB" if there is no anchor and the BES coordinate is within map-derived sequence neighbourhood
- xyy = "ONB" if there is no anchor and the BES coordinate is outside map-derived sequence neighbourhood
- xyy = "DNB" if there is no anchor and the BES coordinate is on a different chromosome as map-derived sequence neighbourhood
- BES--- BES coordinate with no associated fingerprint-based coordinate
- ANC--- fingerprint-based coordinate with no associated BES or Golden Path sequence coordinate
- ------ sequence coordinates could not be determined for this clone
clone covers
This list shows the representation of each part of the assembled sequence by the clones in the set. Each line represents a unique sequence range. The fields are
- chromosome
- start position
- end position
- size of range - 1
- number of rearray BACs in range
- flags
- range annotations
The flags are as follows
- aN - sequence contig gap
- cR - rearray clone
- cRM - rearray clone from fingerprint map
- cM - sequence covered by virtue of map overlap
plate mapping
This list provides the location of all clones in plates. There are more clones in this list than in the coordinates and covers lists because some clones were rejected during fingerprint validation steps, but remain on the plates.
fingerprints
The fingerprints for each clone in the set can be found in this file. Each
fingerprint is represented by a line which contains the mapping clone name,
Genbank name, FPC name, number of fragments, plate and the list of fragments
delimited by ":". The fragment sizes are reported in units of bp.
Fingerprints for -1 is reported for the fragments were had their bands
called manually.
Browsing
UCSC Browser Track
You can browse the clone set with the UCSC Genome Browser, or download the track directly. Clones are coloured by their coordinate type.

Dark clones (1) are localized using available BES coordinates, medium tone clones (2) are localized using fingerprint-based in-silico placement and light clones (3) are placed using their assembly coordinates.
Questions about the clone set should be addressed to Martin Krzywinski (BCGSC)