Proteomics Research
The GSC Proteomics Platform has two primary research areas:
- The functional analysis of gene products (proteins) with aberrant expression in tumours, or identified as somatically mutated through tumour genome sequencing;
- To quantify the global proteome changes in patient tumour samples to identify biomarkers for diagnosis, prognosis, and drug response.
Specialized Techniques
Quantitative Global Proteomics
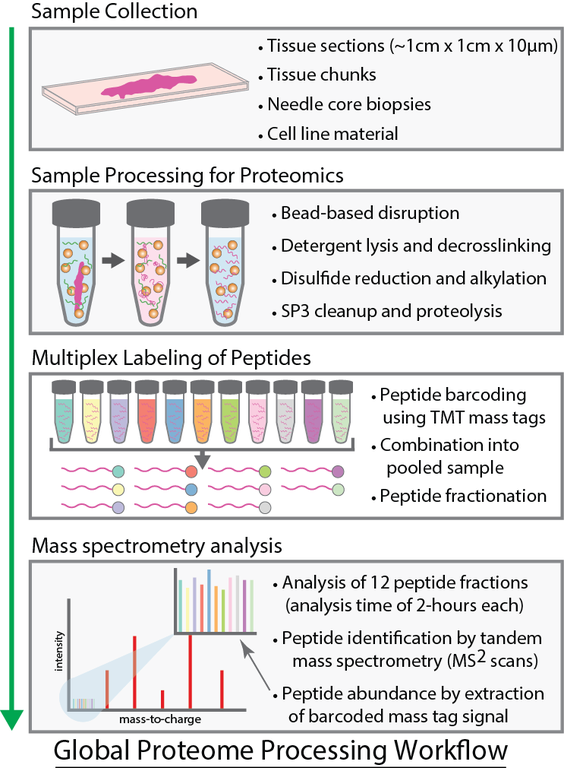
The Proteomics Platform is on the cutting edge of quantitative global proteome analysis. The Tandem Mass Tag (TMT) based discovery proteomics pipeline is used to routinely detect and quantify 8,000+ proteins in a sample and can be applied to whole proteome or immunoprecipitation/affinity purification. Methods have been developed for co-isolation of nucleic acids and proteins from a single homogenous sample as well as surface protein enrichment. Tissues can be processed from formalin-fixed paraffin-embedded (FFPE), DMSO fixed, fresh frozen, or cores. Cells and biological fluids are also amenable to this protocol.
 Targeted Proteomics
Targeted Proteomics
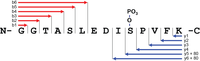
Targeted quantification of proteins, protein complexes, protein pathways, mutated proteins, splice isoforms, and biomarkers using MRM, SRM, and PRM techniques are routinely used for precise and robust quantification of specific proteins. Cells, biological fluids, or tissues (FFPE or DMSO fixed, fresh frozen, or cores) are amenable to this protocol.
Small Molecule Quantification
Targeted methods have been developed for select small molecules, and global metabolomics methods are in development.
Protein Biochemistry
The Platform develops isogenic cell culture systems to study aberrant protein function, and to express and purify recombinant proteins for protein chemistry research. The Platform is broadly equipped with capabilities for protein engineering, biochemical assays, peptide and protein fractionation techniques, ultracentrifugation, and density gradient analyses.
Applications
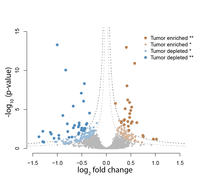
Clinical Tissue ProteomicsThe SP3-CTP (SP3-Clinical Tissue Proteomics) obtains high coverage (>8,000 protein) quantitative proteomics from limited amounts of archived FFPE patient tumour samples1. This transformative technology has led to the discovery of new biomarkers for diverse cancers, and prompted cancer pathologists from BC, Canada, USA, and Europe to initiate projects with the Platform, in order to exploit the potential of their decades-old FFPE tumour sample banks to identify novel cancer biomarkers. |
|
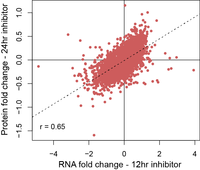
Integrative multi-‘omic profilingThe Platform specializes in the parallel analysis of the genome, transcriptome, and proteome of research and patient samples to understand how genome sequence and RNA processing affect the proteome and cellular processes and biochemistry. |
|
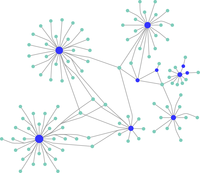
Interactome mappingThe Platform employs TMT methods for the quantitative analysis of protein complexes isolated by chromatographic, affinity purification (direct antibodies or epitope tags), or proximity labeling (BioID, APEX) methods. These data provide direct information on the functions of proteins or mutated proteins in the cell. |
|
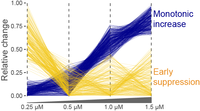
Drug mechanism of actionThe Platform employs TMT methods for the dose and temporal effects of drugs on the global proteome. When coupled with transcriptome information these data provide a comprehensive overview for how drugs or tool compounds affect cellular processes. |
|
Post-Translational Modifications of ProteinsBoth global and targeted methods are implemented for discovery and analysis of post-translational modifications including phosphorylation, ubiquitination, and acetylation. |
|
1Hughes, CS; McConechy, MK; Cochrane, DR; Nazeran, T; Karnezis AN, Huntsman, DG, Morin, GB. Quantitative Profiling of Single Formalin Fixed Tumour Sections: proteomics for translational research. Sci Rep, 2016.